DGEAR-Web Dashboard Manual
Table of Contents
Differential Gene Expression Analysis Resource (DGEAR)
Introduction
Welcome to DGEAR-Web, an intuitive web-based platform for ensemble-based differential gene expression analysis. DGEAR empowers researchers, scientists, and users from varied backgrounds to perform complex DEG (Differentially Expressed Genes) predictions from gene expression data through a user-friendly, GUI-driven environment.
Architecture Overview
DGEAR-Web is designed with the following key principles:
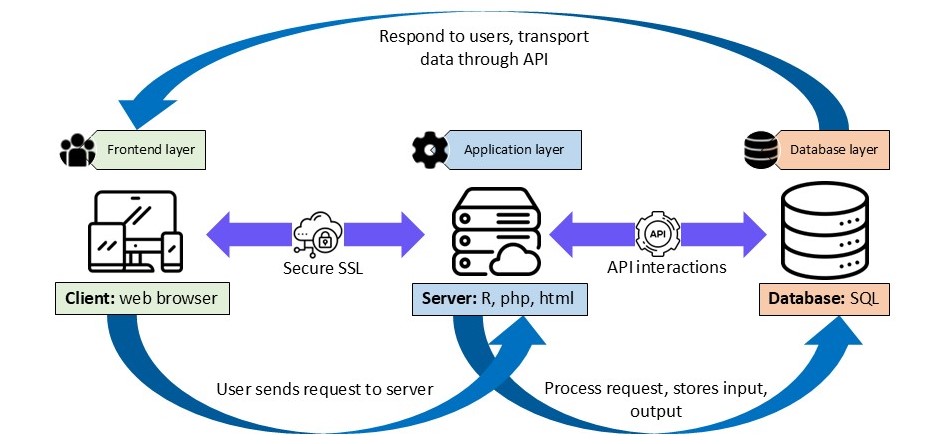
Cross-Platform Compatibility: Accessible across Windows, macOS, Linux, and mobile devices through any modern web browser. User-Friendly Interface: Easy-to-navigate graphical interface with buttons, forms, and menus, ensuring usability without prior programming knowledge. Accessibility: Opens up bioinformatics analysis to users with minimal technical expertise. Efficiency and Time-Saving: Streamlined workflows minimize manual intervention, speeding up the data preprocessing and analysis process.
Key Features
- Upload and analyze gene expression data via simple file upload.
- Customizable input parameters: define control/experimental groups, significance level (alpha), and ensemble voting cutoff.
- Visual outputs: generate plots, results summaries, and downloadable data.
- Result exploration: interactive browsing and download options for outputs.
Data Format and Example Data
- Ensure your data is formatted correctly before upload.
- Example data files are available on the analysis page.
1. Microarray Example Data
| ID | Control1 | Control2 | Experiment1 | Experiment2 |
|---|---|---|---|---|
| A1BG | 4.993423 | 4.977204 | 5.69549 | 5.876676 |
| A1CF | 4.788285 | 4.417162 | 7.557621 | 7.662921 |
| A2M | 4.522844 | 4.679698 | 5.842635 | 5.939148 |
| A2ML1 | 3.864138 | 3.771948 | 4.234489 | 4.342140 |
| A4GALT | 6.248286 | 6.375555 | 7.616046 | 7.489524 |
| ... | ... | ... | ... | ... |
2. RNA-seq Example Data
| GeneID | Control1 | Control2 | Experiment1 | Experiment2 |
|---|---|---|---|---|
| A1BG | 3 | 3 | 3 | 1 |
| A1CF | 367 | 333 | 249 | 277 |
| A2M | 9 | 12 | 4 | 10 |
| A2ML1 | 1 | 1 | 0 | 0 |
| A4GALT | 0 | 0 | 0 | 0 |
| ... | ... | ... | ... | ... |
User Manual
1. Reaching the Web-Tool
Visit https://dgear.compbiosysnbu.in/. Ensure your browser supports JavaScript and cookies for optimal performance. Bookmark the URL for quick access in the future. To download the User Manual, click here!
2. Navigation
Use the navigation panel at the top or bottom. Sections include:
- Home
- Manual
- Analysis
- Results
- Contact
3. Uploading Data
Go to the Analysis tab. Choose Microarray or RNA-seq analysis. Drag and drop or click to upload your file (.tsv, .txt).
4. Setting Input Parameters
After upload, configure: Compare Column Range: e.g., for 4 samples comparing 2 vs 2 → Control: 1–2, Experiment: 3–4 Alpha Value: significance threshold, e.g., 0.05 Voting Cutoff: number of methods that must agree a gene is differentially expressed
- Microarray: 1–5
- RNA-seq: 1–3
5. Submitting and Processing the Request
Click Submit Request. Data will be processed using the ensemble framework.
6. Exploring Results
Once complete, visit the Results Page to:
- View summary plots (e.g., Heatmap, GO bar plots)
- Download DEG lists and outputs from other selected methods.
DGEAR Algorithm
DGEAR implements an ensemble model with a modified majority voting algorithm.
Microarray: Statistical tests: Student’s t-test, ANOVA, Dunnett’s t-test, half t-test, Wilcoxon/Mann-Whitney U-test
RNA-seq: Methods: Linear modeling, negative binomial modeling, empirical Bayes
After FDR correction, results from individual tests are converted to logical vectors, combined via majority voting to determine DEGs.
Important Notes
- Format: genes as rows, samples as columns
- Set input parameters according to your experimental design
- Internet connection is required
Support
For issues or suggestions, contact the project team through the communication options provided on the DGEAR-Web platform.